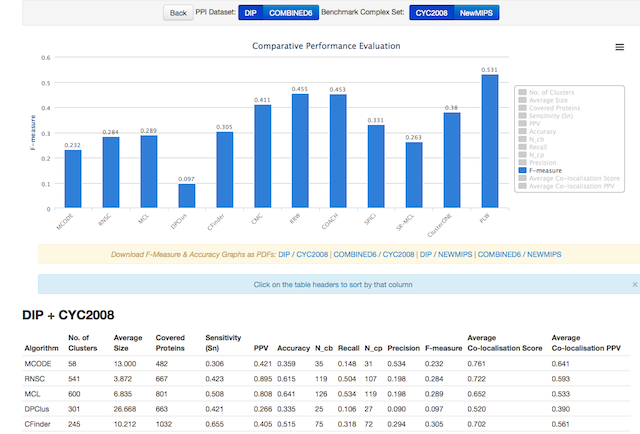
What is PLW?
PLW (Probabilistic Local Walks) is a graph clustering algorithm, and was designed to detect protein complexes in protein-protein interaction (PPI) networks with high accuracy and efficiency.
PLW uses a seed selection strategy to prioritise proteins for expansion into cores, which selects seeds with dense neighbourhoods.
PLW also uses a topological measure called common neighbour similarity to estimate the functional similarity of two proteins in order to weigh the unweighted network. Additionally, you are allowed to supply your own similarity graph in the format of an edge list via a command-line option.
The seed selection and core mining algorithm runs in \(\mathcal{O}(V \log V + E)\) time, where \(V\) is the number of vertices and \(E\) is the number of edges.
Supplementary Materials
Binaries and Datasets
- Binaries and sample data (.zip | .tar.gz)
- Predicted protein complexes (.zip | .tar.gz)
- PPI Datasets Used (.zip | .tar.gz)
- Gold Standard Benchmarks
- CYC2008 Catalogue (Original source: Wodak Lab)
- NewMIPS (Original sources: MIPS, Aloy et al. and SGD Database based on GO annotations)
References
Daniel Lin-Kit Wong, Xiao-Li Li, Min Wu, Jie Zheng and See-Kiong Ng
PLW: Probabilistic Local Walks for detecting protein complexes from protein interaction networks
BMC Genomics 2013 Volume 14 Suppl 5, S15.
http://www.biomedcentral.com/1471-2164/14/S5/S15
Presented in International Conference on Bioinformatics 2013 (InCoB2013).
BiBTeX
@article{Wong2013PLW,
title={PLW: Probabilistic Local Walks for detecting protein complexes from protein interaction networks},
author={Daniel Lin-Kit Wong and Li, Xiao-Li and Wu, Min and Zheng, Jie and Ng, See-Kiong},
journal={BMC Genomics},
doi = {10.1186/1471-2164-14-s5-s15},
url = {http://dx.doi.org/10.1186/1471-2164-14-s5-s15},
volume={14},
number={Suppl 5},
pages={S15},
year={2013},
publisher={BioMed Central Ltd},
pmid = {24564427},
issn = {1471-2164},
abstract = {{BACKGROUND: Many biological processes are carried out by proteins interacting with each other in the form of protein complexes. However, large-scale detection of protein complexes has remained constrained by experimental limitations. As such, computational detection of protein complexes by applying clustering algorithms on the abundantly available protein-protein interaction (PPI) networks is an important alternative. However, many current algorithms have overlooked the importance of selecting seeds for expansion into clusters without excluding important proteins and including many noisy ones, while ensuring a high degree of functional homogeneity amongst the proteins detected for the complexes.
RESULTS: We designed a novel method called Probabilistic Local Walks (PLW) which clusters regions in a PPI network with high functional similarity to find protein complex cores with high precision and efficiency in O(|V| log |V| + |E|) time. A seed selection strategy, which prioritises seeds with dense neighbourhoods, was devised. We defined a topological measure, called common neighbour similarity, to estimate the functional similarity of two proteins given the number of their common neighbours.
CONCLUSIONS: Our proposed PLW algorithm achieved the highest F-measure (recall and precision) when compared to 11 state-of-the-art methods on yeast protein interaction data, with an improvement of 16.7\% over the next highest score. Our experiments also demonstrated that our seed selection strategy is able to increase algorithm precision when applied to three previous protein complex mining techniques.AVAILABILITY:The software, datasets and predicted complexes are available at http://wonglkd.github.io/PLW}}
}
Contact
Websites: Daniel Wong, Dr. Li Xiao-Li, Dr. Wu Min, Dr. Zheng Jie, Dr. Ng See-Kiong
Have a question? Drop me (Daniel) an email at and I will be happy to answer your queries.
Found a bug? Report it on the issue tracker.